Download Survey Package In R
Survey analysis
Overview
This page demonstrates the use of several packages for survey analysis.
Most survey R packages rely on the survey package for doing weighted analysis. We will use survey as well as srvyr (a wrapper for survey allowing for tidyverse-style coding) and gtsummary (a wrapper for survey allowing for publication ready tables). While the original survey package does not allow for tidyverse-style coding, it does have the added benefit of allowing for survey-weighted generalised linear models (which will be added to this page at a later date). We will also demonstrate using a function from the sitrep package to create sampling weights (n.b this package is currently not yet on CRAN, but can be installed from github).
Most of this page is based off work done for the "R4Epis" project; for detailed code and R-markdown templates see the "R4Epis" github page. Some of the survey package based code is based off early versions of EPIET case studies.
At current this page does not address sample size calculations or sampling. For a simple to use sample size calculator see OpenEpi. The GIS basics page of the handbook will eventually have a section on spatial random sampling, and this page will eventually have a section on sampling frames as well as sample size calculations.
- Survey data
- Observation time
- Weighting
- Survey design objects
- Descriptive analysis
- Weighted proportions
- Weighted rates
Preparation
Packages
This code chunk shows the loading of packages required for the analyses. In this handbook we emphasize p_load() from pacman, which installs the package if necessary and loads it for use. You can also load packages with library() from base R. See the page on R basics for more information on R packages.
Here we also demonstrate using the p_load_gh() function from pacman to install a load a package from github which has not yet been published on CRAN.
## load packages from CRAN pacman :: p_load ( rio, # File import here, # File locator tidyverse, # data management + ggplot2 graphics tsibble, # handle time series datasets survey, # for survey functions srvyr, # dplyr wrapper for survey package gtsummary, # wrapper for survey package to produce tables apyramid, # a package dedicated to creating age pyramids patchwork, # for combining ggplots ggforce # for alluvial/sankey plots ) ## load packages from github pacman :: p_load_gh ( "R4EPI/sitrep" # for observation time / weighting functions ) Load data
The example dataset used in this section:
- fictional mortality survey data.
- fictional population counts for the survey area.
- data dictionary for the fictional mortality survey data.
This is based off the MSF OCA ethical review board pre-approved survey. The fictional dataset was produced as part of the "R4Epis" project. This is all based off data collected using KoboToolbox, which is a data collection software based off Open Data Kit.
Kobo allows you to export both the collected data, as well as the data dictionary for that dataset. We strongly recommend doing this as it simplifies data cleaning and is useful for looking up variables/questions.
TIP: The Kobo data dictionary has variable names in the "name" column of the survey sheet. Possible values for each variable are specified in choices sheet. In the choices tab, "name" has the shortened value and the "label::english" and "label::french" columns have the appropriate long versions. Using the epidict package msf_dict_survey() function to import a Kobo dictionary excel file will re-format this for you so it can be used easily to recode.
CAUTION: The example dataset is not the same as an export (as in Kobo you export different questionnaire levels individually) - see the survey data section below to merge the different levels.
The dataset is imported using the import() function from the rio package. See the page on Import and export for various ways to import data.
# import the survey data survey_data <- rio :: import ( "survey_data.xlsx" ) # import the dictionary into R survey_dict <- rio :: import ( "survey_dict.xlsx" ) The first 10 rows of the survey are displayed below.
We also want to import the data on sampling population so that we can produce appropriate weights. This data can be in different formats, however we would suggest to have it as seen below (this can just be typed in to an excel).
# import the population data population <- rio :: import ( "population.xlsx" ) The first 10 rows of the survey are displayed below.
For cluster surveys you may want to add survey weights at the cluster level. You could read this data in as above. Alternatively if there are only a few counts, these could be entered as below in to a tibble. In any case you will need to have one column with a cluster identifier which matches your survey data, and another column with the number of households in each cluster.
## define the number of households in each cluster cluster_counts <- tibble (cluster = c ( "village_1", "village_2", "village_3", "village_4", "village_5", "village_6", "village_7", "village_8", "village_9", "village_10" ), households = c ( 700, 400, 600, 500, 300, 800, 700, 400, 500, 500 ) ) Clean data
The below makes sure that the date column is in the appropriate format. There are several other ways of doing this (see the Working with dates page for details), however using the dictionary to define dates is quick and easy.
We also create an age group variable using the age_categories() function from epikit - see cleaning data handbook section for details. In addition, we create a character variable defining which district the various clusters are in.
Finally, we recode all of the yes/no variables to TRUE/FALSE variables - otherwise these cant be used by the survey proportion functions.
## select the date variable names from the dictionary DATEVARS <- survey_dict %>% filter ( type == "date" ) %>% filter ( name %in% names ( survey_data ) ) %>% ## filter to match the column names of your data pull ( name ) # select date vars ## change to dates survey_data <- survey_data %>% mutate ( across ( all_of ( DATEVARS ), as.Date ) ) ## add those with only age in months to the year variable (divide by twelve) survey_data <- survey_data %>% mutate (age_years = if_else ( is.na ( age_years ), age_months / 12, age_years ) ) ## define age group variable survey_data <- survey_data %>% mutate (age_group = age_categories ( age_years, breakers = c ( 0, 3, 15, 30, 45 ) ) ) ## create a character variable based off groups of a different variable survey_data <- survey_data %>% mutate (health_district = case_when ( cluster_number %in% c ( 1 : 5 ) ~ "district_a", TRUE ~ "district_b" ) ) ## select the yes/no variable names from the dictionary YNVARS <- survey_dict %>% filter ( type == "yn" ) %>% filter ( name %in% names ( survey_data ) ) %>% ## filter to match the column names of your data pull ( name ) # select yn vars ## change to dates survey_data <- survey_data %>% mutate ( across ( all_of ( YNVARS ), str_detect, pattern = "yes" ) ) Survey data
There numerous different sampling designs that can be used for surveys. Here we will demonstrate code for: - Stratified - Cluster - Stratified and cluster
As described above (depending on how you design your questionnaire) the data for each level would be exported as a separate dataset from Kobo. In our example there is one level for households and one level for individuals within those households.
These two levels are linked by a unique identifier. For a Kobo dataset this variable is "_index" at the household level, which matches the "_parent_index" at the individual level. This will create new rows for household with each matching individual, see the handbook section on joining for details.
## join the individual and household data to form a complete data set survey_data <- left_join ( survey_data_hh, survey_data_indiv, by = c ( "_index" = "_parent_index" ) ) ## create a unique identifier by combining indeces of the two levels survey_data <- survey_data %>% mutate (uid = str_glue ( "{index}_{index_y}" ) ) Observation time
For mortality surveys we want to now how long each individual was present for in the location to be able to calculate an appropriate mortality rate for our period of interest. This is not relevant to all surveys, but particularly for mortality surveys this is important as they are conducted frequently among mobile or displaced populations.
To do this we first define our time period of interest, also known as a recall period (i.e. the time that participants are asked to report on when answering questions). We can then use this period to set inappropriate dates to missing, i.e. if deaths are reported from outside the period of interest.
## set the start/end of recall period ## can be changed to date variables from dataset ## (e.g. arrival date & date questionnaire) survey_data <- survey_data %>% mutate (recall_start = as.Date ( "2018-01-01" ), recall_end = as.Date ( "2018-05-01" ) ) # set inappropriate dates to NA based on rules ## e.g. arrivals before start, departures departures after end survey_data <- survey_data %>% mutate ( arrived_date = if_else ( arrived_date < recall_start, as.Date ( NA ), arrived_date ), birthday_date = if_else ( birthday_date < recall_start, as.Date ( NA ), birthday_date ), left_date = if_else ( left_date > recall_end, as.Date ( NA ), left_date ), death_date = if_else ( death_date > recall_end, as.Date ( NA ), death_date ) ) We can then use our date variables to define start and end dates for each individual. We can use the find_start_date() function from sitrep to fine the causes for the dates and then use that to calculate the difference between days (person-time).
start date: Earliest appropriate arrival event within your recall period Either the beginning of your recall period (which you define in advance), or a date after the start of recall if applicable (e.g. arrivals or births)
end date: Earliest appropriate departure event within your recall period Either the end of your recall period, or a date before the end of recall if applicable (e.g. departures, deaths)
## create new variables for start and end dates/causes survey_data <- survey_data %>% ## choose earliest date entered in survey ## from births, household arrivals, and camp arrivals find_start_date ( "birthday_date", "arrived_date", period_start = "recall_start", period_end = "recall_end", datecol = "startdate", datereason = "startcause" ) %>% ## choose earliest date entered in survey ## from camp departures, death and end of the study find_end_date ( "left_date", "death_date", period_start = "recall_start", period_end = "recall_end", datecol = "enddate", datereason = "endcause" ) ## label those that were present at the start/end (except births/deaths) survey_data <- survey_data %>% mutate ( ## fill in start date to be the beginning of recall period (for those empty) startdate = if_else ( is.na ( startdate ), recall_start, startdate ), ## set the start cause to present at start if equal to recall period ## unless it is equal to the birth date startcause = if_else ( startdate == recall_start & startcause != "birthday_date", "Present at start", startcause ), ## fill in end date to be end of recall period (for those empty) enddate = if_else ( is.na ( enddate ), recall_end, enddate ), ## set the end cause to present at end if equall to recall end ## unless it is equal to the death date endcause = if_else ( enddate == recall_end & endcause != "death_date", "Present at end", endcause ) ) ## Define observation time in days survey_data <- survey_data %>% mutate (obstime = as.numeric ( enddate - startdate ) ) Weighting
It is important that you drop erroneous observations before adding survey weights. For example if you have observations with negative observation time, you will need to check those (you can do this with the assert_positive_timespan() function from sitrep. Another thing is if you want to drop empty rows (e.g. with drop_na(uid)) or remove duplicates (see handbook section on De-duplication for details). Those without consent need to be dropped too.
In this example we filter for the cases we want to drop and store them in a separate data frame - this way we can describe those that were excluded from the survey. We then use the anti_join() function from dplyr to remove these dropped cases from our survey data.
DANGER: You cant have missing values in your weight variable, or any of the variables relevant to your survey design (e.g. age, sex, strata or cluster variables).
## store the cases that you drop so you can describe them (e.g. non-consenting ## or wrong village/cluster) dropped <- survey_data %>% filter ( ! consent | is.na ( startdate ) | is.na ( enddate ) | village_name == "other" ) ## use the dropped cases to remove the unused rows from the survey data set survey_data <- anti_join ( survey_data, dropped, by = names ( dropped ) ) As mentioned above we demonstrate how to add weights for three different study designs (stratified, cluster and stratified cluster). These require information on the source population and/or the clusters surveyed. We will use the stratified cluster code for this example, but use whichever is most appropriate for your study design.
# stratified ------------------------------------------------------------------ # create a variable called "surv_weight_strata" # contains weights for each individual - by age group, sex and health district survey_data <- add_weights_strata (x = survey_data, p = population, surv_weight = "surv_weight_strata", surv_weight_ID = "surv_weight_ID_strata", age_group, sex, health_district ) ## cluster --------------------------------------------------------------------- # get the number of people of individuals interviewed per household # adds a variable with counts of the household (parent) index variable survey_data <- survey_data %>% add_count ( index, name = "interviewed" ) ## create cluster weights survey_data <- add_weights_cluster (x = survey_data, cl = cluster_counts, eligible = member_number, interviewed = interviewed, cluster_x = village_name, cluster_cl = cluster, household_x = index, household_cl = households, surv_weight = "surv_weight_cluster", surv_weight_ID = "surv_weight_ID_cluster", ignore_cluster = FALSE, ignore_household = FALSE ) # stratified and cluster ------------------------------------------------------ # create a survey weight for cluster and strata survey_data <- survey_data %>% mutate (surv_weight_cluster_strata = surv_weight_strata * surv_weight_cluster ) Survey design objects
Create survey object according to your study design. Used the same way as data frames to calculate weight proportions etc. Make sure that all necessary variables are created before this.
There are four options, comment out those you do not use: - Simple random - Stratified - Cluster - Stratified cluster
For this template - we will pretend that we cluster surveys in two separate strata (health districts A and B). So to get overall estimates we need have combined cluster and strata weights.
As mentioned previously, there are two packages available for doing this. The classic one is survey and then there is a wrapper package called srvyr that makes tidyverse-friendly objects and functions. We will demonstrate both, but note that most of the code in this chapter will use srvyr based objects. The one exception is that the gtsummary package only accepts survey objects.
Survey package
The survey package effectively uses base R coding, and so it is not possible to use pipes (%>%) or other dplyr syntax. With the survey package we use the svydesign() function to define a survey object with appropriate clusters, weights and strata.
NOTE: we need to use the tilde (~) in front of variables, this is because the package uses the base R syntax of assigning variables based on formulae.
# simple random --------------------------------------------------------------- base_survey_design_simple <- svydesign (ids = ~ 1, # 1 for no cluster ids weights = NULL, # No weight added strata = NULL, # sampling was simple (no strata) data = survey_data # have to specify the dataset ) ## stratified ------------------------------------------------------------------ base_survey_design_strata <- svydesign (ids = ~ 1, # 1 for no cluster ids weights = ~ surv_weight_strata, # weight variable created above strata = ~ health_district, # sampling was stratified by district data = survey_data # have to specify the dataset ) # cluster --------------------------------------------------------------------- base_survey_design_cluster <- svydesign (ids = ~ village_name, # cluster ids weights = ~ surv_weight_cluster, # weight variable created above strata = NULL, # sampling was simple (no strata) data = survey_data # have to specify the dataset ) # stratified cluster ---------------------------------------------------------- base_survey_design <- svydesign (ids = ~ village_name, # cluster ids weights = ~ surv_weight_cluster_strata, # weight variable created above strata = ~ health_district, # sampling was stratified by district data = survey_data # have to specify the dataset ) Srvyr package
With the srvyr package we can use the as_survey_design() function, which has all the same arguments as above but allows pipes (%>%), and so we do not need to use the tilde (~).
## simple random --------------------------------------------------------------- survey_design_simple <- survey_data %>% as_survey_design (ids = 1, # 1 for no cluster ids weights = NULL, # No weight added strata = NULL # sampling was simple (no strata) ) ## stratified ------------------------------------------------------------------ survey_design_strata <- survey_data %>% as_survey_design (ids = 1, # 1 for no cluster ids weights = surv_weight_strata, # weight variable created above strata = health_district # sampling was stratified by district ) ## cluster --------------------------------------------------------------------- survey_design_cluster <- survey_data %>% as_survey_design (ids = village_name, # cluster ids weights = surv_weight_cluster, # weight variable created above strata = NULL # sampling was simple (no strata) ) ## stratified cluster ---------------------------------------------------------- survey_design <- survey_data %>% as_survey_design (ids = village_name, # cluster ids weights = surv_weight_cluster_strata, # weight variable created above strata = health_district # sampling was stratified by district ) Descriptive analysis
Basic descriptive analysis and visualisation is covered extensively in other chapters of the handbook, so we will not dwell on it here. For details see the chapters on descriptive tables, statistical tests, tables for presentation, ggplot basics and R markdown reports.
In this section we will focus on how to investigate bias in your sample and visualise this. We will also look at visualising population flow in a survey setting using alluvial/sankey diagrams.
In general, you should consider including the following descriptive analyses:
- Final number of clusters, households and individuals included
- Number of excluded individuals and the reasons for exclusion
- Median (range) number of households per cluster and individuals per household
Sampling bias
Compare the proportions in each age group between your sample and the source population. This is important to be able to highlight potential sampling bias. You could similarly repeat this looking at distributions by sex.
Note that these p-values are just indicative, and a descriptive discussion (or visualisation with age-pyramids below) of the distributions in your study sample compared to the source population is more important than the binomial test itself. This is because increasing sample size will more often than not lead to differences that may be irrelevant after weighting your data.
## counts and props of the study population ag <- survey_data %>% group_by ( age_group ) %>% drop_na ( age_group ) %>% tally ( ) %>% mutate (proportion = n / sum ( n ), n_total = sum ( n ) ) ## counts and props of the source population propcount <- population %>% group_by ( age_group ) %>% tally ( population ) %>% mutate (proportion = n / sum ( n ) ) ## bind together the columns of two tables, group by age, and perform a ## binomial test to see if n/total is significantly different from population ## proportion. ## suffix here adds to text to the end of columns in each of the two datasets left_join ( ag, propcount, by = "age_group", suffix = c ( "", "_pop" ) ) %>% group_by ( age_group ) %>% ## broom::tidy(binom.test()) makes a data frame out of the binomial test and ## will add the variables p.value, parameter, conf.low, conf.high, method, and ## alternative. We will only use p.value here. You can include other ## columns if you want to report confidence intervals mutate (binom = list ( broom :: tidy ( binom.test ( n, n_total, proportion_pop ) ) ) ) %>% unnest (cols = c ( binom ) ) %>% # important for expanding the binom.test data frame mutate (proportion_pop = proportion_pop * 100 ) %>% ## Adjusting the p-values to correct for false positives ## (because testing multiple age groups). This will only make ## a difference if you have many age categories mutate (p.value = p.adjust ( p.value, method = "holm" ) ) %>% ## Only show p-values over 0.001 (those under report as <0.001) mutate (p.value = ifelse ( p.value < 0.001, "<0.001", as.character ( round ( p.value, 3 ) ) ) ) %>% ## rename the columns appropriately select ( "Age group" = age_group, "Study population (n)" = n, "Study population (%)" = proportion, "Source population (n)" = n_pop, "Source population (%)" = proportion_pop, "P-value" = p.value ) ## # A tibble: 5 x 6 ## # Groups: Age group [5] ## `Age group` `Study population (n)` `Study population (%)` `Source population (n)` `Source population (%)` `P-value` ## <chr> <int> <dbl> <dbl> <dbl> <chr> ## 1 0-2 12 0.0256 1360 6.8 <0.001 ## 2 3-14 42 0.0896 7244 36.2 <0.001 ## 3 15-29 64 0.136 5520 27.6 <0.001 ## 4 30-44 52 0.111 3232 16.2 0.002 ## 5 45+ 299 0.638 2644 13.2 <0.001 Demographic pyramids
Demographic (or age-sex) pyramids are an easy way of visualising the distribution in your survey population. It is also worth considering creating descriptive tables of age and sex by survey strata. We will demonstrate using the apyramid package as it allows for weighted proportions using our survey design object created above. Other options for creating demographic pyramids are covered extensively in that chapter of the handbook. We will also use a wrapper function from sitrep called plot_age_pyramid() which saves a few lines of coding for producing a plot with proportions.
As with the formal binomial test of difference, seen above in the sampling bias section, we are interested here in visualising whether our sampled population is substantially different from the source population and whether weighting corrects this difference. To do this we will use the patchwork package to show our ggplot visualisations side-by-side; for details see the section on combining plots in ggplot tips chapter of the handbook. We will visualise our source population, our un-weighted survey population and our weighted survey population. You may also consider visualising by each strata of your survey - in our example here that would be by using the argument stack_by = "health_district" (see ?plot_age_pyramid for details).
NOTE: The x and y axes are flipped in pyramids
## define x-axis limits and labels --------------------------------------------- ## (update these numbers to be the values for your graph) max_prop <- 35 # choose the highest proportion you want to show step <- 5 # choose the space you want beween labels ## this part defines vector using the above numbers with axis breaks breaks <- c ( seq ( max_prop / 100 * - 1, 0 - step / 100, step / 100 ), 0, seq ( 0 + step / 100, max_prop / 100, step / 100 ) ) ## this part defines vector using the above numbers with axis limits limits <- c ( max_prop / 100 * - 1, max_prop / 100 ) ## this part defines vector using the above numbers with axis labels labels <- c ( seq ( max_prop, step, - step ), 0, seq ( step, max_prop, step ) ) ## create plots individually -------------------------------------------------- ## plot the source population ## nb: this needs to be collapsed for the overall population (i.e. removing health districts) source_population <- population %>% ## ensure that age and sex are factors mutate (age_group = factor ( age_group, levels = c ( "0-2", "3-14", "15-29", "30-44", "45+" ) ), sex = factor ( sex ) ) %>% group_by ( age_group, sex ) %>% ## add the counts for each health district together summarise (population = sum ( population ) ) %>% ## remove the grouping so can calculate overall proportion ungroup ( ) %>% mutate (proportion = population / sum ( population ) ) %>% ## plot pyramid age_pyramid ( age_group = age_group, split_by = sex, count = proportion, proportional = TRUE ) + ## only show the y axis label (otherwise repeated in all three plots) labs (title = "Source population", y = "", x = "Age group (years)" ) + ## make the x axis the same for all plots scale_y_continuous (breaks = breaks, limits = limits, labels = labels ) ## plot the unweighted sample population sample_population <- plot_age_pyramid ( survey_data, age_group = "age_group", split_by = "sex", proportion = TRUE ) + ## only show the x axis label (otherwise repeated in all three plots) labs (title = "Unweighted sample population", y = "Proportion (%)", x = "" ) + ## make the x axis the same for all plots scale_y_continuous (breaks = breaks, limits = limits, labels = labels ) ## plot the weighted sample population weighted_population <- survey_design %>% ## make sure the variables are factors mutate (age_group = factor ( age_group ), sex = factor ( sex ) ) %>% plot_age_pyramid ( age_group = "age_group", split_by = "sex", proportion = TRUE ) + ## only show the x axis label (otherwise repeated in all three plots) labs (title = "Weighted sample population", y = "", x = "" ) + ## make the x axis the same for all plots scale_y_continuous (breaks = breaks, limits = limits, labels = labels ) ## combine all three plots ---------------------------------------------------- ## combine three plots next to eachother using + source_population + sample_population + weighted_population + ## only show one legend and define theme ## note the use of & for combining theme with plot_layout() plot_layout (guides = "collect" ) & theme (legend.position = "bottom", # move legend to bottom legend.title = element_blank ( ), # remove title text = element_text (size = 18 ), # change text size axis.text.x = element_text (angle = 45, hjust = 1, vjust = 1 ) # turn x-axis text ) 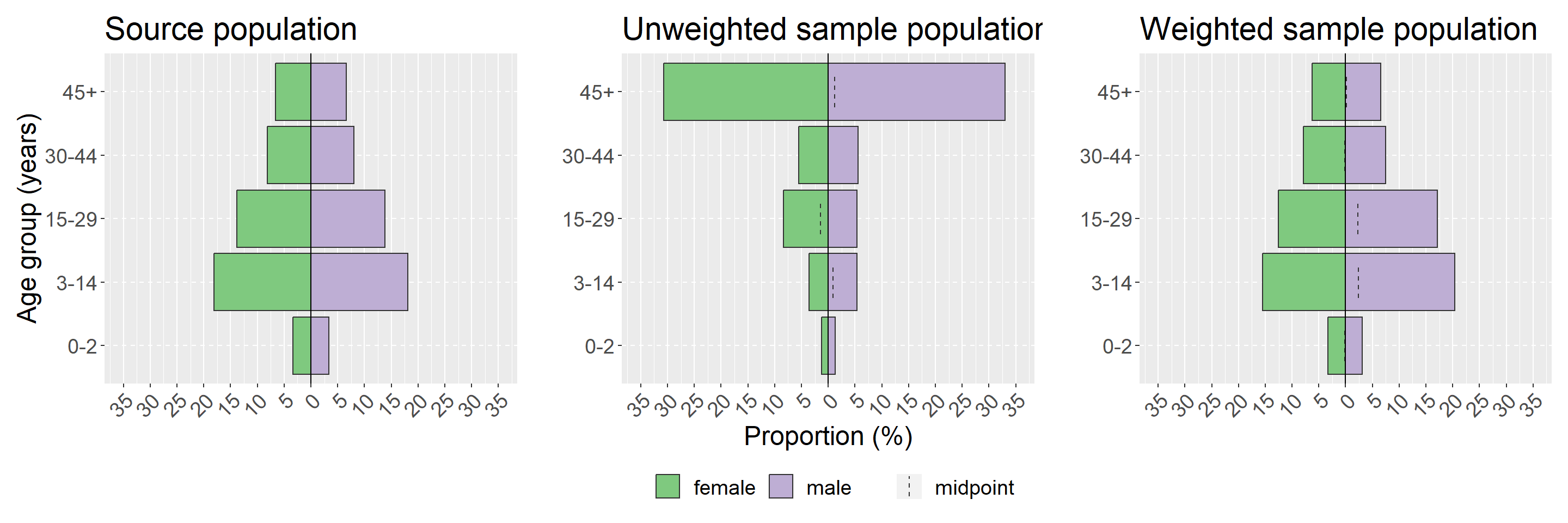
Alluvial/sankey diagram
Visualising starting points and outcomes for individuals can be very helpful to get an overview. There is quite an obvious application for mobile populations, however there are numerous other applications such as cohorts or any other situation where there are transitions in states for individuals. These diagrams have several different names including alluvial, sankey and parallel sets - the details are in the handbook chapter on diagrams and charts.
## summarize data flow_table <- survey_data %>% count ( startcause, endcause, sex ) %>% # get counts gather_set_data (x = c ( "startcause", "endcause" ) ) %>% # change format for plotting mutate (x = fct_relevel ( x, c ( "startcause", "endcause" ) ), # set startcause as first level x = fct_recode ( x, "Start \n cause" = "startcause", # add line break (\n) after start "End \n cause" = "endcause" ) ) ## plot your dataset ## on the x axis is the start and end causes ## gather_set_data generates an ID for each possible combination ## splitting by y gives the possible start/end combos ## value as n gives it as counts (could also be changed to proportion) ggplot ( flow_table, aes ( x, id = id, split = y, value = n ) ) + ## colour lines by sex geom_parallel_sets ( aes (fill = sex ), alpha = 0.5, axis.width = 0.2 ) + ## fill in the label boxes grey geom_parallel_sets_axes (axis.width = 0.15, fill = "grey80", color = "grey80" ) + ## change text colour and angle (needs to be adjusted) geom_parallel_sets_labels (color = "black", angle = 0, size = 5 ) + ## adjusted y and x axes (probably needs more vertical space) scale_x_discrete (name = NULL, expand = c ( 0, 0.2 ) ) + ## remove axis labels theme ( title = element_text (size = 26 ), text = element_text (size = 26 ), axis.line = element_blank ( ), axis.ticks = element_blank ( ), axis.text.y = element_blank ( ), panel.background = element_blank ( ), legend.position = "bottom", # move legend to bottom legend.title = element_blank ( ), # remove title ) 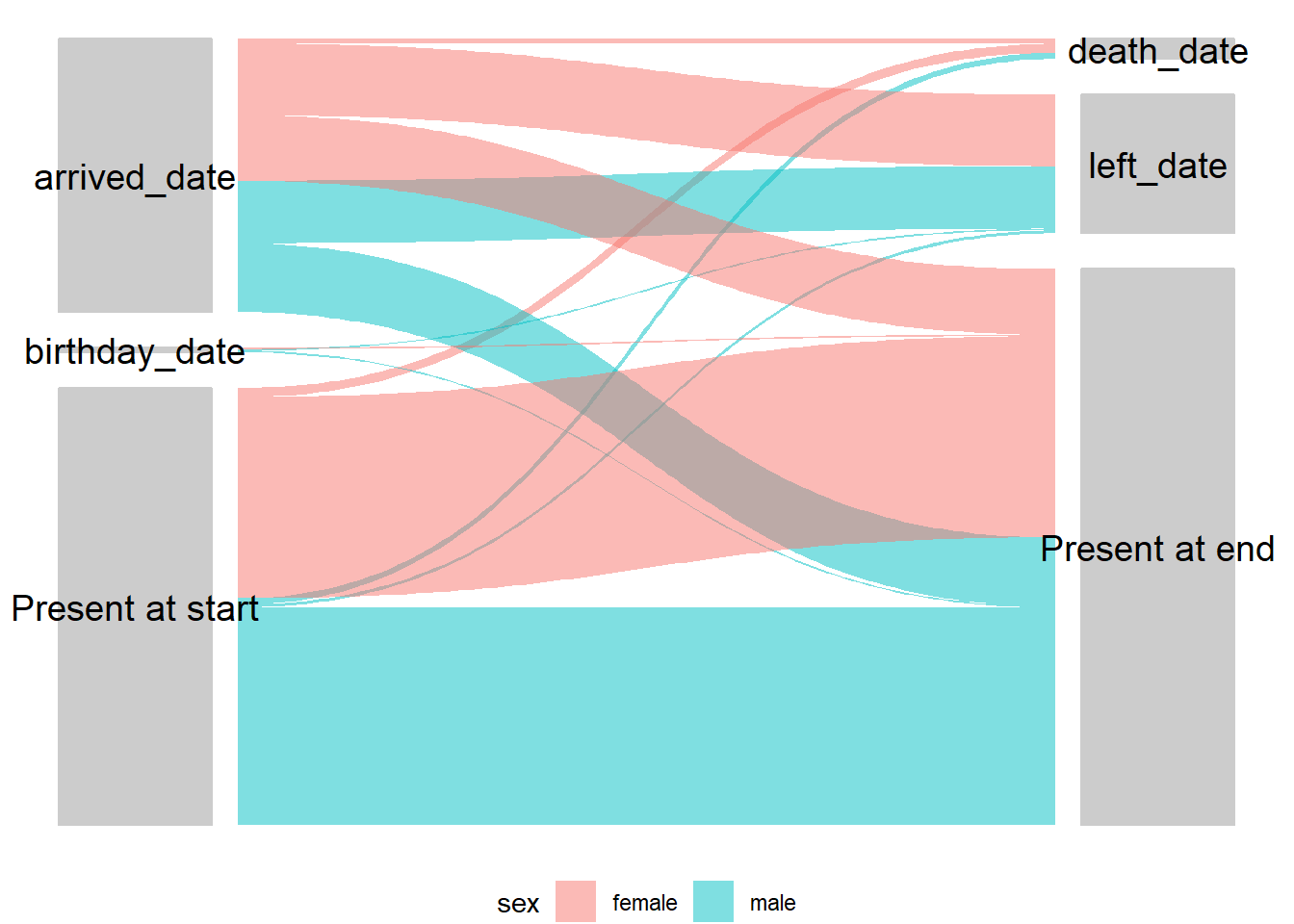
Weighted proportions
This section will detail how to produce tables for weighted counts and proportions, with associated confidence intervals and design effect. There are four different options using functions from the following packages: survey, srvyr, sitrep and gtsummary. For minimal coding to produce a standard epidemiology style table, we would recommend the sitrep function - which is a wrapper for srvyr code; note however that this is not yet on CRAN and may change in the future. Otherwise, the survey code is likely to be the most stable long-term, whereas srvyr will fit most nicely within tidyverse work-flows. While gtsummary functions hold a lot of potential, they appear to be experimental and incomplete at the time of writing.
Survey package
We can use the svyciprop() function from survey to get weighted proportions and accompanying 95% confidence intervals. An appropriate design effect can be extracted using the svymean() rather than svyprop() function. It is worth noting that svyprop() only appears to accept variables between 0 and 1 (or TRUE/FALSE), so categorical variables will not work.
NOTE: Functions from survey also accept srvyr design objects, but here we have used the survey design object just for consistency
## produce weighted counts svytable ( ~ died, base_survey_design ) ## died ## FALSE TRUE ## 1406244.43 76213.01 ## produce weighted proportions svyciprop ( ~ died, base_survey_design, na.rm = T ) ## 2.5% 97.5% ## died 0.0514 0.0208 0.12 ## get the design effect svymean ( ~ died, base_survey_design, na.rm = T, deff = T ) %>% deff ( ) ## diedFALSE diedTRUE ## 3.755508 3.755508 We can combine the functions from survey shown above in to a function which we define ourselves below, called svy_prop; and we can then use that function together with map() from the purrr package to iterate over several variables and create a table. See the handbook iteration chapter for details on purrr.
# Define function to calculate weighted counts, proportions, CI and design effect # x is the variable in quotation marks # design is your survey design object svy_prop <- function ( design, x ) { ## put the variable of interest in a formula form <- as.formula ( paste0 ( "~" , x ) ) ## only keep the TRUE column of counts from svytable weighted_counts <- svytable ( form, design ) [[ 2 ] ] ## calculate proportions (multiply by 100 to get percentages) weighted_props <- svyciprop ( form, design, na.rm = TRUE ) * 100 ## extract the confidence intervals and multiply to get percentages weighted_confint <- confint ( weighted_props ) * 100 ## use svymean to calculate design effect and only keep the TRUE column design_eff <- deff ( svymean ( form, design, na.rm = TRUE, deff = TRUE ) ) [[ TRUE ] ] ## combine in to one data frame full_table <- cbind ( "Variable" = x, "Count" = weighted_counts, "Proportion" = weighted_props, weighted_confint, "Design effect" = design_eff ) ## return table as a dataframe full_table <- data.frame ( full_table, ## remove the variable names from rows (is a separate column now) row.names = NULL ) ## change numerics back to numeric full_table [ , 2 : 6 ] <- as.numeric ( full_table [, 2 : 6 ] ) ## return dataframe full_table } ## iterate over several variables to create a table purrr :: map ( ## define variables of interest c ( "left", "died", "arrived" ), ## state function using and arguments for that function (design) svy_prop, design = base_survey_design ) %>% ## collapse list in to a single data frame bind_rows ( ) %>% ## round mutate ( across ( where ( is.numeric ), round, digits = 1 ) ) ## Variable Count Proportion X2.5. X97.5. Design.effect ## 1 left 701199.1 47.3 39.2 55.5 2.4 ## 2 died 76213.0 5.1 2.1 12.1 3.8 ## 3 arrived 761799.0 51.4 40.9 61.7 3.9 Srvyr package
With srvyr we can use dplyr syntax to create a table. Note that the survey_mean() function is used and the proportion argument is specified, and also that the same function is used to calculate design effect. This is because srvyr wraps around both of the survey package functions svyciprop() and svymean(), which are used in the above section.
NOTE: It does not seem to be possible to get proportions from categorical variables using srvyr either, if you need this then check out the section below using sitrep
## use the srvyr design object survey_design %>% summarise ( ## produce the weighted counts counts = survey_total ( died ), ## produce weighted proportions and confidence intervals ## multiply by 100 to get a percentage props = survey_mean ( died, proportion = TRUE, vartype = "ci" ) * 100, ## produce the design effect deff = survey_mean ( died, deff = TRUE ) ) %>% ## only keep the rows of interest ## (drop standard errors and repeat proportion calculation) select ( counts, props, props_low, props_upp, deff_deff ) ## counts props props_low props_upp deff_deff ## 1 76213.01 5.140991 2.082773 12.13328 3.755508 Here too we could write a function to then iterate over multiple variables using the purrr package. See the handbook iteration chapter for details on purrr.
# Define function to calculate weighted counts, proportions, CI and design effect # design is your survey design object # x is the variable in quotation marks srvyr_prop <- function ( design, x ) { summarise ( ## using the survey design object design, ## produce the weighted counts counts = survey_total ( .data [[ x ] ] ), ## produce weighted proportions and confidence intervals ## multiply by 100 to get a percentage props = survey_mean ( .data [[ x ] ], proportion = TRUE, vartype = "ci" ) * 100, ## produce the design effect deff = survey_mean ( .data [[ x ] ], deff = TRUE ) ) %>% ## add in the variable name mutate (variable = x ) %>% ## only keep the rows of interest ## (drop standard errors and repeat proportion calculation) select ( variable, counts, props, props_low, props_upp, deff_deff ) } ## iterate over several variables to create a table purrr :: map ( ## define variables of interest c ( "left", "died", "arrived" ), ## state function using and arguments for that function (design) ~ srvyr_prop ( .x, design = survey_design ) ) %>% ## collapse list in to a single data frame bind_rows ( ) ## variable counts props props_low props_upp deff_deff ## 1 left 701199.14 47.299782 39.235598 55.50736 2.379761 ## 2 died 76213.01 5.140991 2.082773 12.13328 3.755508 ## 3 arrived 761799.05 51.387583 40.927349 61.72766 3.925504 Sitrep package
The tab_survey() function from sitrep is a wrapper for srvyr, allowing you to create weighted tables with minimal coding. It also allows you to calculate weighted proportions for categorical variables.
## using the survey design object survey_design %>% ## pass the names of variables of interest unquoted tab_survey ( arrived, left, died, education_level, deff = TRUE, # calculate the design effect pretty = TRUE # merge the proportion and 95%CI ) ## Warning: removing 257 missing value(s) from `education_level` ## # A tibble: 9 x 5 ## variable value n deff ci ## <chr> <chr> <dbl> <dbl> <chr> ## 1 arrived TRUE 761799. 3.93 51.4% (40.9--61.7) ## 2 arrived FALSE 720658. 3.93 48.6% (38.3--59.1) ## 3 left TRUE 701199. 2.38 47.3% (39.2--55.5) ## 4 left FALSE 781258. 2.38 52.7% (44.5--60.8) ## 5 died TRUE 76213. 3.76 5.1% (2.1--12.1) ## 6 died FALSE 1406244. 3.76 94.9% (87.9--97.9) ## 7 education_level higher 171644. 4.70 42.4% (26.9--59.7) ## 8 education_level primary 102609. 2.37 25.4% (16.2--37.3) ## 9 education_level secondary 130201. 6.68 32.2% (16.5--53.3) Gtsummary package
With gtsummary there does not seem to be inbuilt functions yet to add confidence intervals or design effect. Here we show how to define a function for adding confidence intervals and then add confidence intervals to a gtsummary table created using the tbl_svysummary() function.
confidence_intervals <- function ( data, variable, by, ... ) { ## extract the confidence intervals and multiply to get percentages props <- svyciprop ( as.formula ( paste0 ( "~" , variable ) ), data, na.rm = TRUE ) ## extract the confidence intervals as.numeric ( confint ( props ) * 100 ) %>% ## make numeric and multiply for percentage round ( ., digits = 1 ) %>% ## round to one digit c ( . ) %>% ## extract the numbers from matrix paste0 ( ., collapse = "-" ) ## combine to single character } ## using the survey package design object tbl_svysummary ( base_survey_design, include = c ( arrived, left, died ), ## define variables want to include statistic = list ( everything ( ) ~ c ( "{n} ({p}%)" ) ) ) %>% ## define stats of interest add_n ( ) %>% ## add the weighted total add_stat (fns = everything ( ) ~ confidence_intervals ) %>% ## add CIs ## modify the column headers modify_header ( list ( n ~ "**Weighted total (N)**", stat_0 ~ "**Weighted Count**", add_stat_1 ~ "**95%CI**" ) ) | Characteristic | Weighted total (N) | Weighted Count 1 | 95%CI |
|---|---|---|---|
| arrived | 1,482,457 | 761,799 (51%) | 40.9-61.7 |
| left | 1,482,457 | 701,199 (47%) | 39.2-55.5 |
| died | 1,482,457 | 76,213 (5.1%) | 2.1-12.1 |
| 1 n (%) | |||
Weighted ratios
Similarly for weighted ratios (such as for mortality ratios) you can use the survey or the srvyr package. You could similarly write functions (similar to those above) to iterate over several variables. You could also create a function for gtsummary as above but currently it does not have inbuilt functionality.
Survey package
ratio <- svyratio ( ~ died, denominator = ~ obstime, design = base_survey_design ) ci <- confint ( ratio ) cbind ( ratio $ ratio * 10000, ci * 10000 ) ## obstime 2.5 % 97.5 % ## died 5.981922 1.194294 10.76955 Srvyr package
survey_design %>% ## survey ratio used to account for observation time summarise ( mortality = survey_ratio ( as.numeric ( died ) * 10000, obstime, vartype = "ci" ) ) ## mortality mortality_low mortality_upp ## 1 5.981922 0.3490176 11.61483 Source: https://epirhandbook.com/en/survey-analysis.html
Posted by: riverbeyond.blogspot.com